Fit an environmental vector or factor to a Co-CA ordination
envfit.coca.RdThe function fits environmental vectors or factors to a Co-CA ordination. The projections of points onto vectors have maximum correlation with corresponding environmental variables, and the factors show the averages of factor levels.
# S3 method for coca envfit(ord, env, which = c("response", "predictor"), choices = c(1, 2), scaling = FALSE, w, na.rm = FALSE, strata = NULL, permutations = 999, ...)
Arguments
| ord | a Co-CA ordination object, the result of a call to
|
|---|---|
| env | a data frame, matrix or vector of environmental/external variable(s) to be fitted to the ordination. The variables may be of a mixed type (factors and continuous variables) in a data frame. |
| which | character; which of the response or predictor ordinations should be used during fitting of vectors and factors. |
| choices | numeric; the axes to which vectors and factors are fitted. |
| scaling | logical; should sacling be applied. See
|
| w | weights used in fitting vectors and factors. |
| na.rm | Remove points with missing values in ordination scores or
environmental variables. The operation is casewise; the whole row of
data is removed if there is a missing value and |
| strata | An integer vector or factor specifying the strata for permutation. If supplied, observations are permuted only within the specified strata. |
| permutations | Number of permutations for assessing significance
of vectors or factors. Set to |
| ... | Arguments passed to |
Details
See envfit for details of the method.
Value
Returns an object of class envfit.
Author
Gavin L. Simpson. The code interfaces with and uses code from
envfit for the main computations, which was written by
Jari Oksanen.
See also
coca for fitting models. envfit for
details of the generic function and the computations performed.
Examples
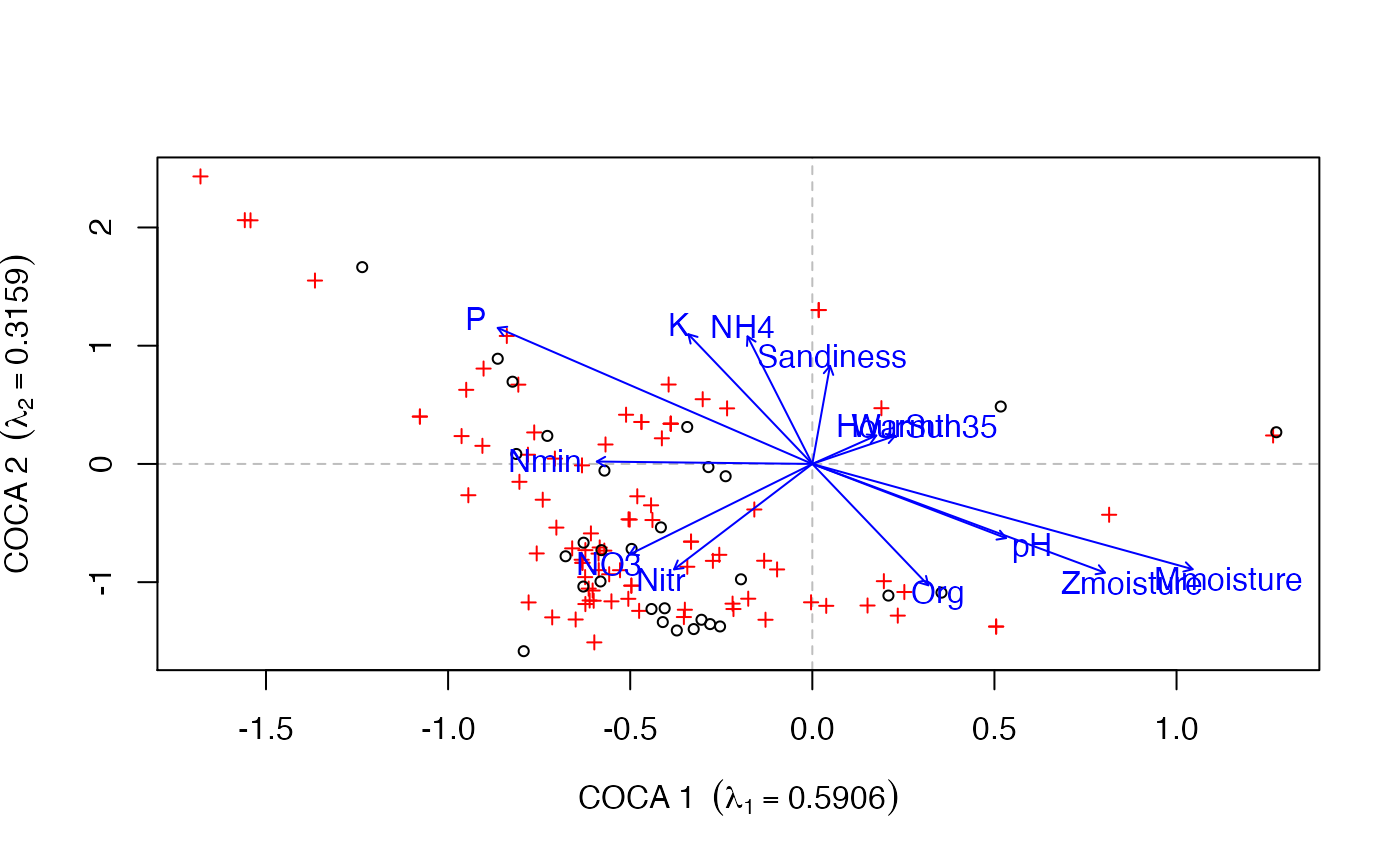
## symmetric CoCA data(beetles) data(plants) ## log transform the bettle data beetles <- log(beetles + 1) ## fit the model bp.sym <- coca(beetles ~ ., data = plants, method = "symmetric") #> #> Removed some species that contained no data in: beetles, plants ## load the environmental data data(verges) ## fit vectors for the environmental data sol <- envfit(bp.sym, verges, which = "response") sol #> #> ***VECTORS #> #> COCA 1 COCA 2 r2 Pr(>r) #> Mmoisture 0.76095 -0.64881 0.4686 0.036 * #> Zmoisture 0.65812 -0.75292 0.3711 0.076 . #> Org 0.29636 -0.95508 0.2873 0.162 #> Sandiness 0.05840 0.99829 0.1718 0.422 #> pH 0.64658 -0.76284 0.1683 0.490 #> NO3 -0.54818 -0.83636 0.2088 0.294 #> NH4 -0.16274 0.98667 0.2975 0.181 #> Nmin -0.99942 0.03412 0.0872 0.678 #> P -0.60030 0.79978 0.5153 0.034 * #> K -0.29634 0.95508 0.3284 0.114 #> Nitr -0.39179 -0.92005 0.2336 0.274 #> HourSun 0.59022 0.80724 0.0220 0.878 #> Warmth35 0.69088 0.72297 0.0269 0.833 #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #> Permutation: free #> Number of permutations: 999 #> #> ## plot the response matrix and the fitted vectors biplot(bp.sym, which = "y1") plot(sol)